Data Analytics | Machine Learning | Classification | ROBERTA | Ensembles
This project was completed as part of the Machine Learning module at SMU. The objective was to train a model which can determine the relationship between sentences. As of 2022, the state-of-the-art method of a determining such relationships was via transformers built on the attention model. Basically by using various queries, keys and values to encode a sentence we are able to extract different types of features be it topological or lexicographical.
The training data is basically a list of sentence pairs, one called an evidence and the other a claim. Each pair is labeled as having a 'supportive', 'refutive' or 'none' relationship. In the transformer model each sentence is encoded and the encodings cross multiplied to create a unique matrix which is then used to train the model as it identifies the matrix as belonging to one of the labels.
The report explaining the project is as seen below. Most of the exploration and writing was completed by me, however the base code that sets up the ROBERTA transformer model for training was adapted from https://github.com/teacherpeterpan/Zero-shot-Fact-Verification.





The contents of the notebook appended below dive straight into the first of 6 stages:
import pandas as pd
claim_evidence =pd.read_csv("train_update.csv")
claim_evidence
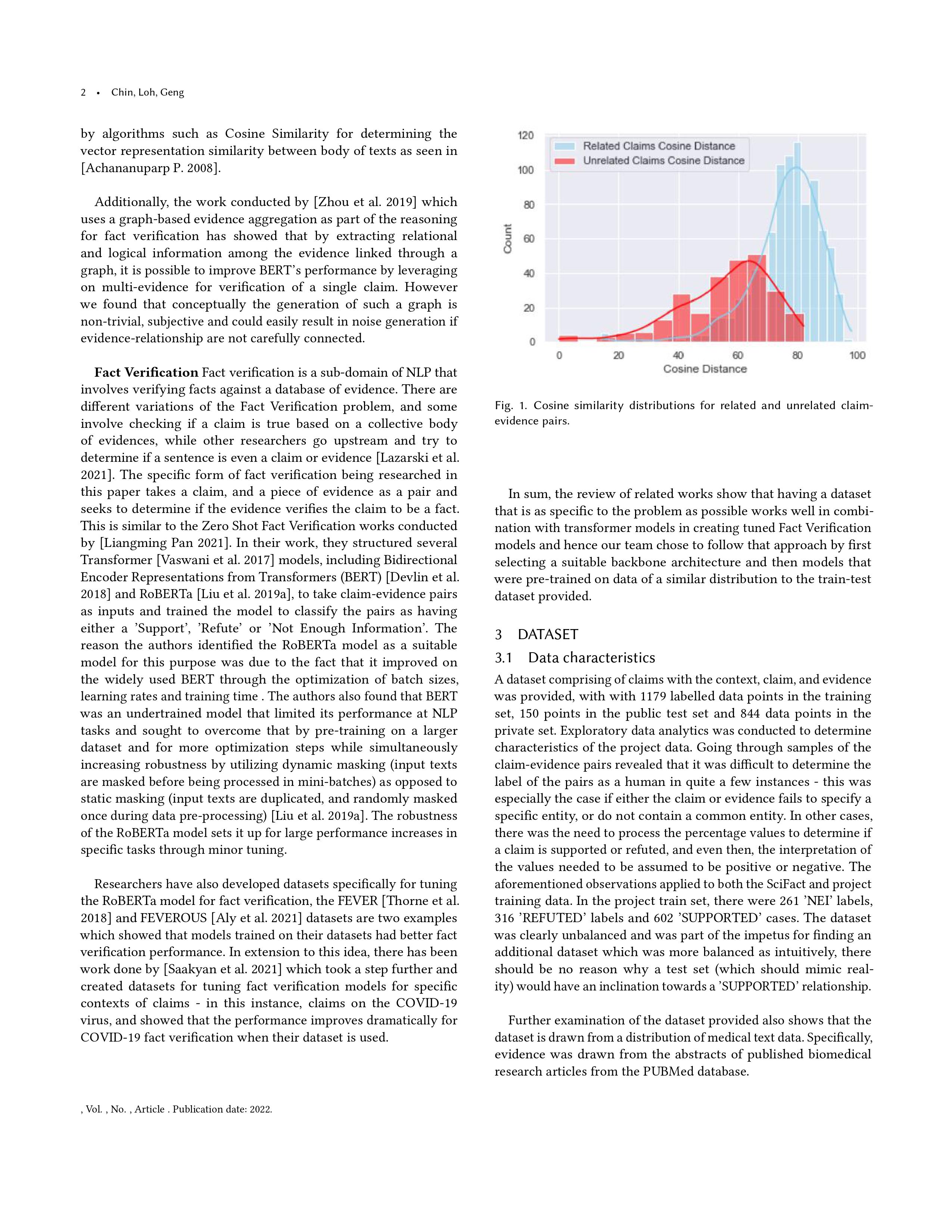
| id | context | claim | evidence_entity | evidence_id | evidence | label | |
|---|---|---|---|---|---|---|---|
| 0 | 0 | Tricyclics were also more likely to reduce the... | Antidepressants increase the severity of migra... | NaN | 0 | Tricyclics were also more likely to reduce the... | REFUTED |
| 1 | 1 | These observations implicate accelerated NETos... | Citrullinated proteins externalized in neutrop... | NaN | 1 | These observations implicate accelerated NETos... | SUPPORTED |
| 2 | 2 | IF3 and tRNA undergo large conformational chan... | Recognition of start codons depends on the tra... | NaN | 2 | IF3 and tRNA undergo large conformational chan... | SUPPORTED |
| 3 | 3 | Conversely, enhanced Ca2+ cycling by activatio... | Ca2+ cycling is a UCP1-dependent thermogenic m... | NaN | 3 | Conversely, enhanced Ca2+ cycling by activatio... | REFUTED |
| 4 | 4 | The Hippo pathway controls organ size and tiss... | Weighed food records (WFR) result in poor comp... | NaN | 4 | The Hippo pathway controls organ size and tiss... | NEI |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 1174 | 1174 | The relative risk estimate for breast cancer c... | Birth-weight is negatively associated with bre... | NaN | 1174 | The relative risk estimate for breast cancer c... | REFUTED |
| 1175 | 1175 | Cry1 expression was elevated during the night-... | Rhythmic expression of Cry1 translates directl... | NaN | 1175 | Cry1 expression was elevated during the night-... | SUPPORTED |
| 1176 | 1176 | Also, mutant mice have increased susceptibilit... | Mice lacking Sirt1 in Sf1-expressing neurons h... | NaN | 1176 | Also, mutant mice have increased susceptibilit... | SUPPORTED |
| 1177 | 1177 | Here, we develop a conceptual framework for fu... | Microglia are an innate immune cell type of th... | NaN | 1177 | Here, we develop a conceptual framework for fu... | REFUTED |
| 1178 | 1178 | The adverse effect is less severe in sons, alt... | There is no increased risk of hypospadias with... | NaN | 1178 | The adverse effect is less severe in sons, alt... | REFUTED |
1179 rows × 7 columns
NEI_claim_evidence = claim_evidence.loc[claim_evidence['label'] == 'NEI']
len(NEI_claim_evidence)
261
REFUTED_claim_evidence = claim_evidence.loc[claim_evidence['label'] == 'REFUTED']
len(REFUTED_claim_evidence)
316
SUPPORTED_claim_evidence = claim_evidence.loc[claim_evidence['label'] == 'SUPPORTED']
len(SUPPORTED_claim_evidence)
602
unrelated_claim_evidence = claim_evidence.loc[claim_evidence['label'] == 'NEI']
related_claim_evidence.reset_index(inplace=True)
related_claim_evidence
| index | id | context | claim | evidence_entity | evidence_id | evidence | label | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | Tricyclics were also more likely to reduce the... | Antidepressants increase the severity of migra... | NaN | 0 | Tricyclics were also more likely to reduce the... | REFUTED |
| 1 | 1 | 1 | These observations implicate accelerated NETos... | Citrullinated proteins externalized in neutrop... | NaN | 1 | These observations implicate accelerated NETos... | SUPPORTED |
| 2 | 2 | 2 | IF3 and tRNA undergo large conformational chan... | Recognition of start codons depends on the tra... | NaN | 2 | IF3 and tRNA undergo large conformational chan... | SUPPORTED |
| 3 | 3 | 3 | Conversely, enhanced Ca2+ cycling by activatio... | Ca2+ cycling is a UCP1-dependent thermogenic m... | NaN | 3 | Conversely, enhanced Ca2+ cycling by activatio... | REFUTED |
| 4 | 5 | 5 | Significantly more patients in the specific ex... | Scapular stabilizer exercises are more effecti... | NaN | 5 | Significantly more patients in the specific ex... | SUPPORTED |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 913 | 1174 | 1174 | The relative risk estimate for breast cancer c... | Birth-weight is negatively associated with bre... | NaN | 1174 | The relative risk estimate for breast cancer c... | REFUTED |
| 914 | 1175 | 1175 | Cry1 expression was elevated during the night-... | Rhythmic expression of Cry1 translates directl... | NaN | 1175 | Cry1 expression was elevated during the night-... | SUPPORTED |
| 915 | 1176 | 1176 | Also, mutant mice have increased susceptibilit... | Mice lacking Sirt1 in Sf1-expressing neurons h... | NaN | 1176 | Also, mutant mice have increased susceptibilit... | SUPPORTED |
| 916 | 1177 | 1177 | Here, we develop a conceptual framework for fu... | Microglia are an innate immune cell type of th... | NaN | 1177 | Here, we develop a conceptual framework for fu... | REFUTED |
| 917 | 1178 | 1178 | The adverse effect is less severe in sons, alt... | There is no increased risk of hypospadias with... | NaN | 1178 | The adverse effect is less severe in sons, alt... | REFUTED |
918 rows × 8 columns
unrelated_claim_evidence.reset_index(inplace=True)
unrelated_claim_evidence
| index | id | context | claim | evidence_entity | evidence_id | evidence | label | |
|---|---|---|---|---|---|---|---|---|
| 0 | 4 | 4 | The Hippo pathway controls organ size and tiss... | Weighed food records (WFR) result in poor comp... | NaN | 4 | The Hippo pathway controls organ size and tiss... | NEI |
| 1 | 11 | 11 | CONCLUSIONS There is a paucity of data regardi... | RA activation of DIF2 and NB4 cells induces ha... | NaN | 11 | CONCLUSIONS There is a paucity of data regardi... | NEI |
| 2 | 14 | 14 | This review examines the use of nanotechnologi... | HNF4A mutations are associated with macrosomia... | NaN | 14 | This review examines the use of nanotechnologi... | NEI |
| 3 | 18 | 18 | Functional brain imaging in three patients sho... | Lamins are found within the inner layer of the... | NaN | 18 | Functional brain imaging in three patients sho... | NEI |
| 4 | 22 | 22 | ['Homeless youth suffer from high rates of hea... | Toll-like receptor (TLR) signaling is involved... | NaN | 22 | ['Homeless youth suffer from high rates of hea... | NEI |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 256 | 1148 | 1148 | In addition, there was an association between ... | Sudden death can occur in patients with orthos... | NaN | 1148 | In addition, there was an association between ... | NEI |
| 257 | 1164 | 1164 | The enzyme which confers resistance to erythro... | In chronic viral infections or tumors, peptide... | NaN | 1164 | The enzyme which confers resistance to erythro... | NEI |
| 258 | 1165 | 1165 | Of those, miR-141 was found to be expressed mo... | Tonic signaling from the scFv is amplified by ... | NaN | 1165 | Of those, miR-141 was found to be expressed mo... | NEI |
| 259 | 1167 | 1167 | Senescence is an irreversible cell-cycle arres... | The artifactual C-terminal helix swapping in t... | NaN | 1167 | Senescence is an irreversible cell-cycle arres... | NEI |
| 260 | 1170 | 1170 | Genome-wide association studies have greatly i... | 1-1% of colorectal cancer patients are diagnos... | NaN | 1170 | Genome-wide association studies have greatly i... | NEI |
261 rows × 8 columns
import scipy
import numpy as np
import re
import nltk
nltk.download('stopwords')
from nltk.corpus import stopwords
import seaborn as sns
import matplotlib.pyplot as plt
gloveFile = "glove.6B.100d.txt"
dimensions = 100
def cosine_distance_countvectorizer_method(s1, s2):
# sentences to list
allsentences = [s1 , s2]
# packages
from sklearn.feature_extraction.text import CountVectorizer
from scipy.spatial import distance
# text to vector
vectorizer = CountVectorizer()
all_sentences_to_vector = vectorizer.fit_transform(allsentences)
text_to_vector_v1 = all_sentences_to_vector.toarray()[0].tolist()
text_to_vector_v2 = all_sentences_to_vector.toarray()[1].tolist()
# distance of similarity
cosine = distance.cosine(text_to_vector_v1, text_to_vector_v2)
print('Similarity of two sentences are equal to ',round((1-cosine)*100,2),'%')
return cosine
def loadGloveModel(gloveFile):
print ("Loading Glove Model")
with open(gloveFile, encoding="utf8" ) as f:
content = f.readlines()
model = {}
for line in content:
splitLine = line.split()
word = splitLine[0]
embedding = np.array([float(val) for val in splitLine[1:]])
model[word] = embedding
print ("Done.",len(model)," words loaded!")
return model
def preprocess(raw_text):
# keep only words
letters_only_text = re.sub("[^a-zA-Z]", " ", raw_text)
# convert to lower case and split
words = letters_only_text.lower().split()
# remove stopwords
stopword_set = set(stopwords.words("english"))
cleaned_words = list(set([w for w in words if w not in stopword_set]))
return cleaned_words
def cosine_distance_between_two_words(word1, word2):
try:
return (1- scipy.spatial.distance.cosine(model[word1], model[word2]))
except:
print('Word distance cannot be computed: ', word1, ' ',word2 )
return 0
def calculate_heat_matrix_for_two_sentences(s1,s2):
s1 = preprocess(s1)
s2 = preprocess(s2)
result_list = [[cosine_distance_between_two_words(word1, word2) for word2 in s2] for word1 in s1]
result_df = pd.DataFrame(result_list)
result_df.columns = s2
result_df.index = s1
return result_df
def cosine_distance_wordembedding_method(s1, s2):
vector1_array = []
vector2_array = []
for word in preprocess(s1):
try:
vector1_array.append(model[word])
except:
vector1_array.append([0 for _ in range(dimensions)])
print(word, " not found in model.")
for word in preprocess(s2):
try:
vector2_array.append(model[word])
except:
vector2_array.append([0 for _ in range(dimensions)])
print(word, " not found in model.")
vector_1 = np.mean(vector1_array,axis=0)
vector_2 = np.mean(vector2_array,axis=0)
#vector_1 = np.mean([model[word] for word in preprocess(s1)],axis=0)
#vector_2 = np.mean([model[word] for word in preprocess(s2)],axis=0)
cosine = scipy.spatial.distance.cosine(vector_1, vector_2)
print('The claim is: ',s1)
print('The evidence is: ',s2)
print('Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to',round((1-cosine)*100,2),'%')
return(round((1-cosine)*100,2))
def heat_map_matrix_between_two_sentences(s1,s2):
df = calculate_heat_matrix_for_two_sentences(s1,s2)
fig, ax = plt.subplots(figsize=(5,5))
ax_blue = sns.heatmap(df, cmap="YlGnBu")
# ax_red = sns.heatmap(df)
print(cosine_distance_wordembedding_method(s1, s2))
return ax_blue
model = loadGloveModel(gloveFile)
Loading Glove Model Done. 400001 words loaded!
import random
random_index = random.choice([num for num in range(len(related_claim_evidence))])
print("Random index selected is: ",random_index)
heat_map_matrix_between_two_sentences(related_claim_evidence['claim'][random_index],related_claim_evidence['evidence'][random_index])
print(cosine_distance_wordembedding_method(related_claim_evidence['claim'][random_index],related_claim_evidence['evidence'][random_index]))
Random index selected is: 101 The claim is: The risk of female prisoners harming themselves is ten times that of male prisoners. The evidence is: FINDINGS 139,195 self-harm incidents were recorded in 26,510 individual prisoners between 2004 and 2009; 5-6% of male prisoners and 20-24% of female inmates self-harmed every year. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 93.48 % 93.48 The claim is: The risk of female prisoners harming themselves is ten times that of male prisoners. The evidence is: FINDINGS 139,195 self-harm incidents were recorded in 26,510 individual prisoners between 2004 and 2009; 5-6% of male prisoners and 20-24% of female inmates self-harmed every year. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 93.48 % 93.48
related_claim_evidence['Cosine Distance'] = 0
for df_entry in range(len(related_claim_evidence)):
related_claim_evidence['Cosine Distance'][df_entry] = cosine_distance_wordembedding_method(related_claim_evidence['Claim'][df_entry],related_claim_evidence['Evidence'][df_entry])
The claim is: Antidepressants increase the severity of migraines. The evidence is: Tricyclics were also more likely to reduce the intensity of headaches by at least 50% than either placebo (tension-type: relative risk 1.41, 95% confidence interval 1.02 to 1.89; migraine: 1.80, 1.24 to 2.62) or selective serotonin reuptake inhibitors (1.73, 1.34 to 2.22 and 1.72, 1.15 to 2.55). Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 77.97 % citrullinated not found in model. netosis not found in model. citrullinated not found in model. immunostimulatory not found in model. autoantigens not found in model. The claim is: Citrullinated proteins externalized in neutrophil extracellular traps act indirectly to perpetuate the inflammatory cycle via induction of autoantibodies. The evidence is: These observations implicate accelerated NETosis in RA pathogenesis, through externalization of citrullinated autoantigens and immunostimulatory molecules that may promote aberrant adaptive and innate immune responses in the joint and in the periphery, and perpetuate pathogenic mechanisms in this disease. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 76.13 % fmet not found in model. formylmethionyl not found in model. The claim is: Recognition of start codons depends on the translation initiation factor IF3. The evidence is: IF3 and tRNA undergo large conformational changes to facilitate the accommodation of the formylmethionyl-tRNA (fMet-tRNA(fMet)) into the P site for start codon recognition. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 78.13 % The claim is: Ca2+ cycling is a UCP1-dependent thermogenic mechanism. The evidence is: Conversely, enhanced Ca2+ cycling by activation of α1- and/or β3-adrenergic receptors or the SERCA2b-RyR2 pathway stimulates UCP1-independent thermogenesis in beige adipocytes. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 60.85 % The claim is: Scapular stabilizer exercises are more effective than general exercise therapy in reducing pain and improving function of the shoulder. The evidence is: Significantly more patients in the specific exercise group reported successful outcome (defined as large improvement or recovered) in the patients' global assessment of change because of treatment: 69% (35/51) v 24% (11/46); odds ratio 7.6, 3.1 to 18.9; P<0.001. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 77.92 % The claim is: Human embryonic stem cells give rise to cell types from the outer embryonic germ layer, but not the other two layers. The evidence is: Embryonic stem cells have the ability to remain undifferentiated and proliferate indefinitely in vitro while maintaining the potential to differentiate into derivatives of all three embryonic germ layers. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 90.66 % hepatocytic not found in model. ihepscs not found in model. hepatocytic not found in model. cholangiocytic not found in model. The claim is: Induced hepatic stem cells possess the potential to differentiate into hepatocytic cells. The evidence is: iHepSCs can be stably expanded in vitro and possess the potential of bidirectional differentiation into both hepatocytic and cholangiocytic lineages. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 76.63 % The claim is: Knockout proximal tubule-specific deletion of the BMP receptor Alk3 causes epithelial damage. The evidence is: Combining THR-123 and the angiotensin-converting enzyme inhibitor captopril had an additive therapeutic benefit in controlling renal fibrosis. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 68.57 % coatmer not found in model. atgl not found in model. ...
related_claim_evidence['Cosine Distance'].describe()
count 918.000000 mean 76.047930 std 11.980168 min 14.000000 25% 70.250000 50% 78.000000 75% 84.000000 max 98.000000 Name: Cosine Distance, dtype: float64
ax = related_claim_evidence['Cosine Distance'].plot.hist(bins=20, alpha=0.5)
import random
random_index = random.choice([num for num in range(len(unrelated_claim_evidence))])
heat_map_matrix_between_two_sentences(unrelated_claim_evidence['Claim'][random_index],unrelated_claim_evidence['Evidence'][random_index])
Word distance cannot be computed: episomally sumo Word distance cannot be computed: episomally ligase Word distance cannot be computed: episomally mms Word distance cannot be computed: episomally docking Word distance cannot be computed: episomally complex Word distance cannot be computed: episomally active Word distance cannot be computed: episomally indicating Word distance cannot be computed: episomally disjunction Word distance cannot be computed: episomally depend Word distance cannot be computed: episomally functions Word distance cannot be computed: episomally intact Word distance cannot be computed: episomally smc Word distance cannot be computed: episomally large Word distance cannot be computed: episomally operates Word distance cannot be computed: episomally chromosome Word distance cannot be computed: episomally vivo Word distance cannot be computed: episomally show Word distance cannot be computed: reational sumo Word distance cannot be computed: reational ligase Word distance cannot be computed: reational mms Word distance cannot be computed: reational docking Word distance cannot be computed: reational complex Word distance cannot be computed: reational active Word distance cannot be computed: reational indicating Word distance cannot be computed: reational disjunction Word distance cannot be computed: reational depend Word distance cannot be computed: reational functions Word distance cannot be computed: reational intact Word distance cannot be computed: reational smc Word distance cannot be computed: reational large Word distance cannot be computed: reational operates Word distance cannot be computed: reational chromosome Word distance cannot be computed: reational vivo Word distance cannot be computed: reational show episomally not found in model. reational not found in model. The claim is: Understanding epigenetic regulation of replication is essential for the reational design of episomally replicating vectors. The evidence is: Here we show that the SUMO ligase and the chromosome disjunction functions of Mms21 depend on its docking to an intact and active Smc5/6 complex, indicating that the Smc5/6-Mms21 complex operates as a large SUMO ligase in vivo. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 67.3 % None
<AxesSubplot:>
unrelated_claim_evidence['Cosine Distance'] = 0
for df_entry in range(len(unrelated_claim_evidence)):
unrelated_claim_evidence['Cosine Distance'][df_entry] = cosine_distance_wordembedding_method(unrelated_claim_evidence['Claim'][df_entry],unrelated_claim_evidence['Evidence'][df_entry])
The claim is: Weighed food records (WFR) result in poor completion since they're costly to run and impose high participant burden. The evidence is: The Hippo pathway controls organ size and tissue homeostasis, with deregulation leading to cancer. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 63.52 % The claim is: RA activation of DIF2 and NB4 cells induces hallmarks of transcriptionally active promoters. The evidence is: CONCLUSIONS There is a paucity of data regarding newborn care-seeking behaviors; in South Asia, care seeking is low for newborn illness, especially in terms of care sought from health care facilities and medically trained providers. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 28.95 % macrosomia not found in model. hnf not found in model. The claim is: HNF4A mutations are associated with macrosomia in infancy. The evidence is: This review examines the use of nanotechnologies for stem cell tracking, differentiation, and transplantation. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 51.06 % The claim is: Lamins are found within the inner layer of the mitochondrial membrane of all cells. The evidence is: Functional brain imaging in three patients showed a focal reduction in hemispheric perfusion most prominent in the ipsilateral inferior and medial frontal cortex. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 70.74 % The claim is: Toll-like receptor (TLR) signaling is involved in the pathogenesis of human MDS. The evidence is: ['Homeless youth suffer from high rates of health problems, yet little is known about their perceptions of or context for their own health issues.'] Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 61.37 % passaging not found in model. The claim is: Pure neural progenitor cell (NPC) populations can only be obtained from cell cultures that undergo passaging, filtration, or other isolation and cell sorting methods. The evidence is: After an initial healing period, the control limbs regenerated normally. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 73.3 % The claim is: British female students are bullied more than British male students. The evidence is: We then discuss the concepts behind the identification of common variants as disease causal factors and contrast them to the basic ideas that underlie the rare variant hypothesis. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 49.32 % hhv not found in model. The claim is: Continued HHV-8 transmission among MSM in San Francisco may be explained by urogenital contact. The evidence is: In the case of guinea worm infection, disease eradication might also soon be possible [9]. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 76.47 % The claim is: Long chain polyunsaturated fatty acids supplementation has no significant effects on wheezing or asthma at 3 and 6 years. The evidence is: In vitro, apolipoprotein E in cerebrospinal fluid binds to synthetic beta A4 peptide (the primary constituent of the senile plaque) with high avidity. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 60.89 % The claim is: Individuals with low serum vitamin D concentrations have decreased risk of multiple sclerosis. The evidence is: We hypothesized that LIF-responsive genes in Shp-2(Delta46-110) cells would represent potential candidates for molecules vital for ES cell self-renewal. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 67.51 % The claim is: NR5A2 is important in reverse cholesterol transport in humans. The evidence is: Current UK and EU legislation limits the number of low strength verbal descriptors and the associated alcohol by volume (ABV) to 1.2% ABV and lower. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 67.69 % The claim is: BiP is a general endoplasmic reticulum stress marker. The evidence is: ['Within each of two study arms, animals were randomized into 5 groups (NPC, SAP, NPC+SAP, vehicle, and sham).'] Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 32.87 % tmao not found in model. The claim is: Sudden death can occur in patients with complex ventricular arrhythmias. The evidence is: Plasma L-carnitine levels in subjects undergoing cardiac evaluation (n = 2,595) predicted increased risks for both prevalent cardiovascular disease (CVD) and incident major adverse cardiac events (myocardial infarction, stroke or death), but only among subjects with concurrently high TMAO levels. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 80.55 % dialyzable not found in model. hdle not found in model. The claim is: Certain immunomodulator-human dialyzable leukocyte extract (hDLE) peptides are recognized by toll-like receptors (TLRs) on macrophages and dendritic cells. The evidence is: Certain pathogens, such as Mycobacterium tuberculosis, survive within the hostile intracellular environment of a macrophage. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 69.75 % tertile not found in model. The claim is: Long - range chromatin interactions regulate transcription. The evidence is: RESULTS After adjustment for age and sex, a low ABI was associated with lower scoring (bottom tertile vs top tertile) on Raven's Matrices (odds ratio (OR)=1.6, 95% confidence interval (CI) =1.0-2.6), Verbal Fluency (OR =1.8, 95% CI =1.1-3.0), and Digit Symbol Test (OR =2.3, 95% CI =1.3-4.2), suggesting that the ABI is predictive of poorer performance in nonverbal reasoning, verbal fluency, and information processing speed. Word Embedding method with a cosine distance asses that this claim and evidence pair are similar to 60.11 % The claim is: 90% of sudden infant death syndrome (SIDS) deaths happen in newborns aged less than 6 months. The evidence is: In animal models, obese mice with fatty livers are vulnerable to liver adenosine triphosphate (ATP) depletion and necrosis, suggesting that altered hepatic energy homeostasis may be involved. ...
unrelated_claim_evidence['Cosine Distance'].describe()
count 261.000000 mean 55.816092 std 15.830979 min 0.000000 25% 46.000000 50% 60.000000 75% 67.000000 max 82.000000 Name: Cosine Distance, dtype: float64
ax = unrelated_claim_evidence['Cosine Distance'].plot.hist(bins=20, alpha=0.5)
#Compare both distributions
# libraries & dataset
import seaborn as sns
import matplotlib.pyplot as plt
# set a grey background (use sns.set_theme() if seaborn version 0.11.0 or above)
sns.set(style="darkgrid")
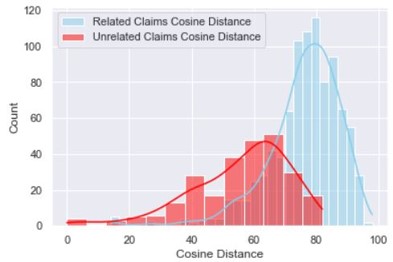
sns.histplot(data=related_claim_evidence, x="Cosine Distance", color="skyblue", label="Related Claims Cosine Distance", kde=True)
sns.histplot(data=unrelated_claim_evidence, x="Cosine Distance", color="red", label="Unrelated Claims Cosine Distance", kde=True)
plt.legend()
plt.show()
Intention is to extract entity names from claims and evidence and bypass overall similarity checks when there are similar entities in both claims and evidence.
import spacy
from spacy import displacy
NER = spacy.load("en_core_web_lg")
import random
random_index = random.choice([num for num in range(len(related_claim_evidence))])
print("Random index selected is: ",random_index)
claim = related_claim_evidence['Claim'][random_index]
evidence = related_claim_evidence['Evidence'][random_index]
entities = NER(claim)
for entity in entities.ents:
print(entity.text,entity.label_)
print(related_claim_evidence['Claim'][random_index])
import spacy
from spacy import displacy
NER = spacy.load("en_core_sci_sm")
import random
random_index = random.choice([num for num in range(len(related_claim_evidence))])
print("Random index selected is: ",random_index)
claim = related_claim_evidence['Claim'][random_index]
evidence = related_claim_evidence['Evidence'][random_index]
entities = NER(claim)
for entity in entities.ents:
print(entity.text,entity.label_)
print(related_claim_evidence['Claim'][random_index])
Random index selected is: 293 Citrullinated proteins ENTITY neutrophil ENTITY extracellular traps ENTITY inflammatory cycle ENTITY induction ENTITY autoantibodies ENTITY Citrullinated proteins externalized in neutrophil extracellular traps act indirectly to perpetuate the inflammatory cycle via induction of autoantibodies.
import spacy
from spacy import displacy
NER = spacy.load("en_core_sci_lg")
import random
random_index = random.choice([num for num in range(len(related_claim_evidence))])
print("Random index selected is: ",random_index)
claim = related_claim_evidence['claim'][random_index]
evidence = related_claim_evidence['evidence'][random_index]
entities = NER(claim)
for entity in entities.ents:
print(entity.text,entity.label_)
print(related_claim_evidence['claim'][random_index])
Random index selected is: 116 Birth-weight ENTITY associated with ENTITY breast cancer ENTITY Birth-weight is negatively associated with breast cancer.
import spacy
from spacy import displacy
NER = spacy.load("en_core_sci_scibert")
import random
random_index = random.choice([num for num in range(len(related_claim_evidence))])
print("Random index selected is: ",random_index)
claim = related_claim_evidence['claim'][random_index]
evidence = related_claim_evidence['evidence'][random_index]
entities = NER(claim)
for entity in entities.ents:
print(entity.text,entity.label_,entity.class_)
print(related_claim_evidence['claim'][random_index])
Random index selected is: 49
import spacy
from spacy import displacy
NER = spacy.load("en_core_sci_lg")
# Default as no match, state set to 0
related_claim_evidence['Entity Match'] = 0
for df_entry in range(len(related_claim_evidence)):
claim = related_claim_evidence['Claim'][df_entry]
claim_entities = NER(claim).ents
split_claim_entities = []
for claim_entity in claim_entities:
for split_entity in str(claim_entity).split():
split_claim_entities.append(split_entity.lower())
evidence = related_claim_evidence['Evidence'][df_entry]
evidence_entities = NER(evidence).ents
split_evidence_entities = []
for evidence_entity in evidence_entities:
for split_entity in str(evidence_entity).split():
split_evidence_entities.append(split_entity.lower())
for entity in split_claim_entities:
if entity in split_evidence_entities:
related_claim_evidence['Entity Match'][df_entry] = 1
# print('claim: ',claim)
# print('evidence: ',evidence)
# print('matched entity: ',entity)
related_claim_evidence['Entity Match'].describe()
count 918.000000 mean 0.844227 std 0.362838 min 0.000000 25% 1.000000 50% 1.000000 75% 1.000000 max 1.000000 Name: Entity Match, dtype: float64
import spacy
from spacy import displacy
NER = spacy.load("en_core_sci_lg")
# Default as no match, state set to 0
unrelated_claim_evidence['Entity Match'] = 0
for df_entry in range(len(unrelated_claim_evidence)):
claim = unrelated_claim_evidence['Claim'][df_entry]
claim_entities = NER(claim).ents
split_claim_entities = []
for claim_entity in claim_entities:
for split_entity in str(claim_entity).split():
split_claim_entities.append(split_entity.lower())
evidence = unrelated_claim_evidence['Evidence'][df_entry]
evidence_entities = NER(evidence).ents
split_evidence_entities = []
for evidence_entity in evidence_entities:
for split_entity in str(evidence_entity).split():
split_evidence_entities.append(split_entity.lower())
for entity in split_claim_entities:
if entity in split_evidence_entities:
unrelated_claim_evidence['Entity Match'][df_entry] = 1
break
# print('claim: ',claim)
# print('evidence: ',evidence)
# print('matched entity: ',entity)
unrelated_claim_evidence['Entity Match'].describe()
count 261.000000 mean 0.045977 std 0.209838 min 0.000000 25% 0.000000 50% 0.000000 75% 0.000000 max 1.000000 Name: Entity Match, dtype: float64
# Implement the sentence similarity check for a sentence made up of just the extracted entities for a non-binary threshold to determine if it is matched or not
There could be words or character combinations (e.g. chemical compounds) that are not in the embeddings, in such cases we need a way to remove non-entity words from the sentence and check for the entity names in both the claim and evidence
There should be a smarter way (than just looking for a word match) for checking entities between claim and evidence to determine if it is a match or not. Fuzzy matching for each word pair? Or just apply the Cosine Similarity on a merge of all the entities?
python run_hover.py --model_type roberta --model_name_or_path roberta-large --do_train --do_lower_case --per_gpu_train_batch_size 16 --learning_rate 1e-5 --num_train_epochs 5.0 --evaluate_during_training --max_seq_length 200 --max_query_length 60 --gradient_accumulation_steps 2 --max_steps 60 --save_steps 60 --logging_steps 60 --overwrite_cache --num_labels 3 --data_dir ../data/ --train_file project_train_data.json --predict_file test_phase_1_update.json --output_dir ./output/roberta_zero_shot
python run_hover.py --model_type roberta --model_name_or_path roberta-large --do_eval --do_lower_case --per_gpu_train_batch_size 16 --learning_rate 1e-5 --num_train_epochs 5.0 --evaluate_during_training --max_seq_length 200 --max_query_length 60 --gradient_accumulation_steps 2 --max_steps 20000 --save_steps 1000 --logging_steps 1000 --overwrite_cache --num_labels 3 --data_dir ../data/ --train_file fever_train_data.json --predict_file test_phase_1_update_human.json --output_dir ./tuned_model_lr1e5_bs16_s75(5)/roberta_zero_shot
python run_hover.py --model_type roberta --model_name_or_path roberta-large --do_eval --do_lower_case --per_gpu_train_batch_size 16 --learning_rate 1e-5 --num_train_epochs 5.0 --evaluate_during_training --max_seq_length 200 --max_query_length 60 --gradient_accumulation_steps 2 --max_steps 20000 --save_steps 1000 --logging_steps 1000 --overwrite_cache --num_labels 3 --data_dir ../data/ --train_file fever_train_data.json --predict_file test_phase_2_update.json --output_dir ./scifact_model3(91.33)/roberta_zero_shot
python run_hover.py --model_type roberta --model_name_or_path roberta-large --do_eval --do_lower_case --per_gpu_train_batch_size 16 --learning_rate 1e-5 --num_train_epochs 5.0 --evaluate_during_training --max_seq_length 200 --max_query_length 60 --gradient_accumulation_steps 2 --max_steps 20000 --save_steps 1000 --logging_steps 1000 --overwrite_cache --num_labels 3 --data_dir ../data/ --train_file fever_train_data.json --predict_file test_phase_1_update_human2.json --output_dir ./tuned_model_lr1e5_bs16_s75(5)/roberta_zero_shot
python run_hover.py --model_type roberta --model_name_or_path ./tuned_model_lr1e5_bs16_s75(5)/roberta_zero_shot/best_model --do_train --do_lower_case --per_gpu_train_batch_size 16 --learning_rate 1e-5 --num_train_epochs 5.0 --evaluate_during_training --max_seq_length 200 --max_query_length 60 --gradient_accumulation_steps 2 --max_steps 100 --save_steps 100 --logging_steps 100 --overwrite_cache --num_labels 3 --data_dir ../data/ --train_file scifact_train_dev.json --predict_file test_phase_1_update_human_2.json --output_dir ./output/roberta_zero_shot
python run_hover.py --model_type roberta --model_name_or_path ./sample_model_3/roberta_zero_shot/best_model --do_train --do_lower_case --per_gpu_train_batch_size 16 --learning_rate 1e-5 --num_train_epochs 5.0 --evaluate_during_training --max_seq_length 200 --max_query_length 60 --gradient_accumulation_steps 2 --max_steps 1000 --save_steps 100 --logging_steps 100 --overwrite_cache --num_labels 3 --data_dir ../data/ --train_file scifact_train_dev_sampled.json --predict_file test_phase_1_update_human_2.json --output_dir ./output/roberta_zero_shot
python run_hover2.py --model_type bert --model_name_or_path microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext --do_train --do_lower_case --per_gpu_train_batch_size 16 --learning_rate 1e-5 --num_train_epochs 5.0 --evaluate_during_training --max_seq_length 200 --max_query_length 60 --gradient_accumulation_steps 1 --save_steps 60 --logging_steps 60 --overwrite_cache --num_labels 3 --data_dir ../data/ --train_file scifact_train_dev_sampled.json --predict_file scifact_train_dev.json --output_dir ./output/roberta_zero_shot
python run_hover2.py --model_type bert --model_name_or_path microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext --do_eval --do_lower_case --per_gpu_train_batch_size 16 --learning_rate 1e-5 --num_train_epochs 5.0 --evaluate_during_training --max_seq_length 200 --max_query_length 60 --gradient_accumulation_steps 1 --save_steps 60 --logging_steps 60 --overwrite_cache --num_labels 3 --data_dir ../data/ --train_file scifact_train_dev_sampled.json --predict_file test_phase_2_update.json --output_dir ./pubmed_tuned_model_40ep_bs16_lr1e5/roberta_zero_shot
filename = 'test_phase_2_update'
import pandas as pd
df = pd.read_csv(filename+'.csv')
import spacy
from spacy import displacy
NER = spacy.load("en_core_sci_lg")
# Default as no match, state set to 0
df['evidence_entity'] = None
for df_entry in range(len(df)):
claim = df['claim'][df_entry]
claim_entities = NER(claim).ents
df['evidence_entity'][df_entry] = claim_entities[0][0]
df['ori_evidence'] = None
for df_entry in range(len(df)):
df['ori_evidence'][df_entry] = [[df['evidence_entity'][df_entry],df['evidence_id'][df_entry],df['evidence'][df_entry]]]
df_selected = df[['id','context','ori_evidence','claim','label']]
records = df_selected.to_json(orient="records",default_handler=str,path_or_buf='./{}.json'.format(filename),indent=1)
Based on absence of shared entity presence in claim-evidence. Forced labels are stored in column 'related'. Amongst the related claim-evidence there is still a need to differentiate between support and refute.
filename = 'test_phase_1_update'
import pandas as pd
df_related = pd.read_csv(filename+'.csv')
import spacy
from spacy import displacy
NER = spacy.load("en_core_sci_lg")
df_related['related'] = 0
for df_entry in range(len(df_related)):
claim = df_related['claim'][df_entry]
claim_entities = NER(claim).ents
split_claim_entities = []
for claim_entity in claim_entities:
for split_entity in str(claim_entity).split():
split_claim_entities.append(split_entity.lower())
evidence = df_related['evidence'][df_entry]
evidence_entities = NER(evidence).ents
split_evidence_entities = []
for evidence_entity in evidence_entities:
for split_entity in str(evidence_entity).split():
split_evidence_entities.append(split_entity.lower())
for entity in split_claim_entities:
if entity in split_evidence_entities:
df_related['related'][df_entry] = 1
break
df_related
| id | context | claim | evidence_entity | evidence_id | evidence | label | related | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | This result suggests that traces of a heavy me... | Beta-band coherence is diminished for visible ... | NaN | 0 | This result suggests that traces of a heavy me... | NEI | 0 |
| 1 | 1 | The best diagnostic performance is obtained wh... | Physical activity level is associated with the... | NaN | 1 | The best diagnostic performance is obtained wh... | NEI | 0 |
| 2 | 2 | After 6 months, 4 of 53 patients (7.5%) in the... | Autologous transplantation of mesenchymal stem... | NaN | 2 | After 6 months, 4 of 53 patients (7.5%) in the... | NEI | 1 |
| 3 | 3 | At CFSs, this fragility is associated with a d... | RAD52 is involved in break-induced DNA replica... | NaN | 3 | At CFSs, this fragility is associated with a d... | NEI | 1 |
| 4 | 4 | Embryonic stem cells have the ability to remai... | Human embryonic stem cells give rise to cell t... | NaN | 4 | Embryonic stem cells have the ability to remai... | NEI | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 145 | 145 | However, mature size DNA products accumulated ... | Origin gross domestic product(GDP) is positive... | NaN | 145 | However, mature size DNA products accumulated ... | NEI | 0 |
| 146 | 146 | RESULTS There was no significant mean treatmen... | Surgical treatment is not superior to non-surg... | NaN | 146 | RESULTS There was no significant mean treatmen... | NEI | 1 |
| 147 | 147 | Further, we demonstrated that LXR is poly(ADP-... | Cancer-associated fibroblasts (CAFs) interact ... | NaN | 147 | Further, we demonstrated that LXR is poly(ADP-... | NEI | 0 |
| 148 | 148 | CONTEXT The epidemic of heart failure has yet ... | Persistor cells are one reason for incomplete ... | NaN | 148 | CONTEXT The epidemic of heart failure has yet ... | NEI | 0 |
| 149 | 149 | Notably, a network structure analysis of this ... | Normal expression of RUNX1 causes tumorsupress... | NaN | 149 | Notably, a network structure analysis of this ... | NEI | 1 |
150 rows × 8 columns
unrelated_claim_evidence_labels
0 0
1 0
2 1
3 1
4 1
..
145 0
146 1
147 0
148 0
149 1
Name: related, Length: 150, dtype: int64
import pandas as pd
import json
import numpy as np
with open('predictions_test_phase_1_update_human_2.json', 'r') as f:
data = json.load(f)
df = pd.DataFrame(data).transpose()
df['submission'] = None
df['submission'] = df['gold_label'].map({'NEI': 0, 'SUPPORTED': 1, 'REFUTED':2})
np.savetxt(r'submission.txt', df.submission, fmt='%s')
import pandas as pd
df = pd.read_csv('logit_results(91.33,4NEI).csv')
filename = 'test_phase_1_update'
import pandas as pd
df_related = pd.read_csv(filename+'.csv')
import spacy
from spacy import displacy
NER = spacy.load("en_core_sci_lg")
df_related['related'] = 0
for df_entry in range(len(df_related)):
claim = df_related['claim'][df_entry]
claim_entities = NER(claim).ents
split_claim_entities = []
for claim_entity in claim_entities:
for split_entity in str(claim_entity).split():
split_claim_entities.append(split_entity.lower())
evidence = df_related['evidence'][df_entry]
evidence_entities = NER(evidence).ents
split_evidence_entities = []
for evidence_entity in evidence_entities:
for split_entity in str(evidence_entity).split():
split_evidence_entities.append(split_entity.lower())
for entity in split_claim_entities:
if entity in split_evidence_entities:
df_related['related'][df_entry] = 1
break
df['related'] = df_related['related'][df_entry]
df
| Unnamed: 0 | id | logit | related | |
|---|---|---|---|---|
| 0 | 0 | 1000000000 | [-0.2988024950027466, -1.1718432903289795, 0.9... | 1 |
| 1 | 1 | 1000000001 | [-1.637555718421936, -1.964380145072937, 3.513... | 1 |
| 2 | 2 | 1000000002 | [1.150705099105835, 1.0391881465911865, -2.863... | 1 |
| 3 | 3 | 1000000003 | [3.090744972229004, -1.1045386791229248, -1.98... | 1 |
| 4 | 4 | 1000000004 | [3.428008556365967, -0.7349969744682312, -2.57... | 1 |
| ... | ... | ... | ... | ... |
| 145 | 145 | 1000000145 | [-0.9186779260635376, -1.5854158401489258, 2.2... | 1 |
| 146 | 146 | 1000000146 | [2.027890205383301, 0.9760094285011292, -3.329... | 1 |
| 147 | 147 | 1000000147 | [-0.22157752513885498, -2.5750298500061035, 2.... | 1 |
| 148 | 148 | 1000000148 | [-1.3043547868728638, -1.696756362915039, 2.83... | 1 |
| 149 | 149 | 1000000149 | [3.1722331047058105, -0.6526208519935608, -2.4... | 1 |
150 rows × 4 columns
# import these modules
import nltk
nltk.download('wordnet')
from nltk.stem import WordNetLemmatizer
lemmatizer = WordNetLemmatizer()
print("rocks :", lemmatizer.lemmatize("rockr"))
print("rocks :", lemmatizer.lemmatize("rocked"))
print("rocks :", lemmatizer.lemmatize("rocks"))
print("corpora :", lemmatizer.lemmatize("corpora"))
# a denotes adjective in "pos"
print("better :", lemmatizer.lemmatize("better", pos ="a"))
rocks : rockr rocks : rocked rocks : rock corpora : corpus better : good
[nltk_data] Downloading package wordnet to [nltk_data] C:\Users\jooer\AppData\Roaming\nltk_data... [nltk_data] Package wordnet is already up-to-date!
import argparse
import jsonlines
from sklearn.feature_extraction.text import TfidfVectorizer
import numpy as np
import pandas as pd
corpus = {doc['doc_id']: doc for doc in jsonlines.open("corpus.jsonl")}
abstract_retrieval = jsonlines.open("abstract_retrieval_train.jsonl")
dataset = jsonlines.open("claims_train.jsonl")
output = jsonlines.open("output.jsonl", 'w')
index = 0
indices = []
context = []
ori_evi = []
claim = []
labels = []
for data, retrieval in zip(dataset, abstract_retrieval):
assert data['id'] == retrieval['claim_id']
for doc_id in retrieval['doc_ids']:
if data['evidence'].get(str(doc_id)):
evidence[doc_id] = [s for es in data['evidence'][str(doc_id)] for s in es['sentences']]
else:
sentences = corpus[doc_id]['abstract']
vectorizer = TfidfVectorizer(stop_words='english')
sentence_vectors = vectorizer.fit_transform(sentences)
claim_vector = vectorizer.transform([data['claim']]).todense()
sentence_scores = np.asarray(sentence_vectors @ claim_vector.T).squeeze()
top_sentence_indices = sentence_scores.argsort()[-2:][::-1].tolist()
top_sentence_indices.sort()
evidence[doc_id] = top_sentence_indices
for sentence_id in evidence[doc_id]:
evidence_sentence = corpus[doc_id]
print(data['evidence'])
try:
label = data['evidence'][str(doc_id)][0]['label']
except:
label = 'NEI'
indices.append(index)
context.append(data['claim'])
ori_evi.append(['','',evidence_sentence['abstract'][sentence_id]])
claim.append(data['claim'])
print(label)
labels.append(label)
print(labels)
output.write({
'id':index,
'claim': data['claim'],
'evidence': evidence_sentence['abstract'][sentence_id],
'label': label})
index += 1
df = pd.DataFrame({'id':indices,'context':context,'ori_evidence':ori_evi,'claim':claim,'label':labels})
...
['NEI', 'NEI', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT']
{'16120395': [{'sentences': [1, 2], 'label': 'SUPPORT'}, {'sentences': [3], 'label': 'SUPPORT'}, {'sentences': [4], 'label': 'SUPPORT'}]}
...
IOPub data rate exceeded. The notebook server will temporarily stop sending output to the client in order to avoid crashing it. To change this limit, set the config variable `--NotebookApp.iopub_data_rate_limit`. Current values: NotebookApp.iopub_data_rate_limit=1000000.0 (bytes/sec) NotebookApp.rate_limit_window=3.0 (secs)
df.replace({'label': {'SUPPORT': 'SUPPORTED', 'CONTRADICT': 'REFUTED'}},inplace=True)
records = df.to_json(orient="records",default_handler=str,path_or_buf='./{}.json'.format('scifact_train'),indent=1)
import argparse
import jsonlines
from sklearn.feature_extraction.text import TfidfVectorizer
import numpy as np
import pandas as pd
corpus = {doc['doc_id']: doc for doc in jsonlines.open("corpus.jsonl")}
abstract_retrieval = jsonlines.open("abstract_retrieval_dev.jsonl")
dataset = jsonlines.open("claims_dev.jsonl")
output = jsonlines.open("output.jsonl", 'w')
index = 0
indices = []
context = []
ori_evi = []
claim = []
labels = []
for data, retrieval in zip(dataset, abstract_retrieval):
assert data['id'] == retrieval['claim_id']
for doc_id in retrieval['doc_ids']:
if data['evidence'].get(str(doc_id)):
evidence[doc_id] = [s for es in data['evidence'][str(doc_id)] for s in es['sentences']]
else:
sentences = corpus[doc_id]['abstract']
vectorizer = TfidfVectorizer(stop_words='english')
sentence_vectors = vectorizer.fit_transform(sentences)
claim_vector = vectorizer.transform([data['claim']]).todense()
sentence_scores = np.asarray(sentence_vectors @ claim_vector.T).squeeze()
top_sentence_indices = sentence_scores.argsort()[-2:][::-1].tolist()
top_sentence_indices.sort()
evidence[doc_id] = top_sentence_indices
for sentence_id in evidence[doc_id]:
evidence_sentence = corpus[doc_id]
print(data['evidence'])
try:
label = data['evidence'][str(doc_id)][0]['label']
except:
label = 'NEI'
indices.append(index)
context.append(data['claim'])
ori_evi.append(['','',evidence_sentence['abstract'][sentence_id]])
claim.append(data['claim'])
print(label)
labels.append(label)
print(labels)
output.write({
'id':index,
'claim': data['claim'],
'evidence': evidence_sentence['abstract'][sentence_id],
'label': label})
index += 1
df2 = pd.DataFrame({'id':indices,'context':context,'ori_evidence':ori_evi,'claim':claim,'label':labels})
df2.replace({'label': {'SUPPORT': 'SUPPORTED', 'CONTRADICT': 'REFUTED'}},inplace=True)
records = df2.to_json(orient="records",default_handler=str,path_or_buf='./{}.json'.format('scifact_dev'),indent=1)
...
['NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'CONTRADICT', 'NEI', 'NEI', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'NEI', 'NEI', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'CONTRADICT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'CONTRADICT', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'NEI', 'NEI', 'NEI', 'NEI', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'SUPPORT', 'CONTRADICT', 'CONTRADICT', 'SUPPORT', 'SUPPORT']
{'16322674': [{'sentences': [5], 'label': 'SUPPORT'}, {'sentences': [6], 'label': 'SUPPORT'}], '27123743': [{'sentences': [3], 'label': 'SUPPORT'}, {'sentences': [4], 'label': 'SUPPORT'}], '23557241': [{'sentences': [6], 'label': 'SUPPORT'}], '17450673': [{'sentences': [5], 'label': 'SUPPORT'}]}
SUPPORT
...
df3 = pd.concat([df,df2])
df3
| id | context | ori_evidence | claim | label | |
|---|---|---|---|---|---|
| 0 | 0 | 0-dimensional biomaterials lack inductive prop... | [, , This review examines the use of nanotechn... | 0-dimensional biomaterials lack inductive prop... | NEI |
| 1 | 1 | 0-dimensional biomaterials lack inductive prop... | [, , We further discuss their utility and the ... | 0-dimensional biomaterials lack inductive prop... | NEI |
| 2 | 2 | 1 in 5 million in UK have abnormal PrP positiv... | [, , RESULTS Of the 32,441 appendix samples 16... | 1 in 5 million in UK have abnormal PrP positiv... | REFUTED |
| 3 | 3 | 1-1% of colorectal cancer patients are diagnos... | [, , During the study period, 44,924 eligible ... | 1-1% of colorectal cancer patients are diagnos... | NEI |
| 4 | 4 | 1-1% of colorectal cancer patients are diagnos... | [, , CONCLUSIONS Expansion of Medicare reimbur... | 1-1% of colorectal cancer patients are diagnos... | NEI |
| ... | ... | ... | ... | ... | ... |
| 585 | 585 | cSMAC formation enhances weak ligand signalling. | [, , This conclusion was supported by experime... | cSMAC formation enhances weak ligand signalling. | SUPPORTED |
| 586 | 586 | mTORC2 regulates intracellular cysteine levels... | [, , mTORC2 phosphorylates serine 26 at the cy... | mTORC2 regulates intracellular cysteine levels... | SUPPORTED |
| 587 | 587 | mTORC2 regulates intracellular cysteine levels... | [, , Genetic inhibition of mTORC2, or pharmaco... | mTORC2 regulates intracellular cysteine levels... | SUPPORTED |
| 588 | 588 | p16INK4A accumulation is linked to an abnorma... | [, , Expression of p16(INK4a) with aging did n... | p16INK4A accumulation is linked to an abnorma... | NEI |
| 589 | 589 | p16INK4A accumulation is linked to an abnorma... | [, , This work suggests that p16(INK4a) activa... | p16INK4A accumulation is linked to an abnorma... | NEI |
2223 rows × 5 columns
df3['id'] = [index for index in range(len(df3))]
records = df3.to_json(orient="records",default_handler=str,path_or_buf='./{}.json'.format('scifact_train_dev'),indent=1)
sample_df = df3.groupby("label").sample(n=480, random_state=1)
print(len(df3.loc[df3['label']=='SUPPORTED']),len(df3.loc[df3['label']=='NEI']),len(df3.loc[df3['label']=='REFUTED']))
896 832 495
print(len(sample_df.loc[sample_df['label']=='SUPPORTED']),len(sample_df.loc[sample_df['label']=='NEI']),len(sample_df.loc[sample_df['label']=='REFUTED']))
records = sample_df.to_json(orient="records",default_handler=str,path_or_buf='./{}.json'.format('scifact_train_dev_sampled'),indent=1)
480 480 480
import pandas as pd
import json
import numpy as np
with open('predictions_test_phase_1_update_human_2.json', 'r') as f:
data = json.load(f)
df_gold = pd.DataFrame(data).transpose()
df_gold['submission'] = None
df_gold['submission'] = df_gold['gold_label'].map({'NEI': 0, 'SUPPORTED': 1, 'REFUTED':2})
df_gold['submission']
0 0
1 0
2 1
3 1
4 1
..
145 0
146 1
147 0
148 0
149 1
Name: submission, Length: 150, dtype: int64
import pandas as pd
logit1_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/tuned_model_lr1e5_bs16_s75(5)/roberta_zero_shot/best_model/logit_results.csv')
logit2_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/scifact_model3(91.33)/roberta_zero_shot/best_model/logit_results.csv')
print(len(logit1_df))
print(len(logit2_df))
150 150
import numpy as np
import ast
ensemble_logit = []
original_logit = []
for logit_tuple1,logit_tuple2 in zip(logit1_df['logit'],logit2_df['logit']):
logit_tuple1 = ast.literal_eval(logit_tuple1)
logit_tuple2 = ast.literal_eval(logit_tuple2)
ensemble_logit.append([float(logit_tuple1[0])+float(logit_tuple2[0]),float(logit_tuple1[1])+float(logit_tuple2[1]),float(logit_tuple1[2])+float(logit_tuple2[2])])
original_logit.append([float(logit_tuple1[0]),float(logit_tuple1[1]),float(logit_tuple1[2])])
print(len(ensemble_logit))
ensemble_logit
150
[[-2.090417742729187, -3.2918331623077393, 4.600583434104919], [-3.3832743167877197, -3.955903172492981, 7.1647772789001465], [1.2023823857307434, 1.742479145526886, -4.153828859329224], ...]
submission = []
original = []
for logits in ensemble_logit:
submission.append(logits.index(max(logits)))
for logits in original_logit:
original.append(logits.index(max(logits)))
changes = 0
ori_error = 0
ensem_error = 0
for entry in range(len(submission)):
if submission[entry] == 0:
submission[entry] = 1
elif submission[entry] == 1:
submission[entry] = 2
elif submission[entry] == 2:
submission[entry] = 0
for entry in range(len(original)):
if original[entry] == 0:
original[entry] = 1
elif original[entry] == 1:
original[entry] = 2
elif original[entry] == 2:
original[entry] = 0
for ori,ensem,gold in zip(original,submission,df_gold['submission']):
print(ori,ensem,gold)
if ori != ensem:
changes += 1
if ori != gold:
ori_error += 1
if ensem != gold:
ensem_error += 1
print(changes)
print(ori_error)
print(ensem_error)
0 0 0 0 0 0 1 2 1 1 1 1 ... 9 15 10
10/150
0.06666666666666667
import pandas as pd
logit1_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/tuned_model_lr1e5_bs16_s75(5)/roberta_zero_shot/best_model/logit_results.csv')
logit2_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/scifact_model3(91.33)/roberta_zero_shot/best_model/logit_results.csv')
logit3_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/scifact_model5/roberta_zero_shot/best_model/logit_results.csv')
print(len(logit1_df))
print(len(logit2_df))
print(len(logit3_df))
150 150 150
import numpy as np
import ast
ensemble_logit = []
original_logit = []
for logit_tuple1,logit_tuple2,logit_tuple3 in zip(logit1_df['logit'],logit2_df['logit'],logit3_df['logit']):
logit_tuple1 = ast.literal_eval(logit_tuple1)
logit_tuple2 = ast.literal_eval(logit_tuple2)
logit_tuple3 = ast.literal_eval(logit_tuple3)
ensemble_logit.append([float(logit_tuple1[0])+float(logit_tuple2[0])+float(logit_tuple3[0]),float(logit_tuple1[1])+float(logit_tuple2[1])+float(logit_tuple3[1]),float(logit_tuple1[2])+float(logit_tuple2[2])+float(logit_tuple3[2])])
original_logit.append([float(logit_tuple1[0]),float(logit_tuple1[1]),float(logit_tuple1[2])])
print(len(ensemble_logit))
ensemble_logit
150
[[-5.003809571266174, -6.264508962631226, 11.532247424125671], [-6.364320755004883, -6.929993748664856, 14.146795272827148], [6.984847366809845, -0.845904529094696, -6.779565095901489], ...]
submission = []
original = []
for logits in ensemble_logit:
submission.append(logits.index(max(logits)))
for logits in original_logit:
original.append(logits.index(max(logits)))
changes = 0
ori_error = 0
ensem_error = 0
for entry in range(len(submission)):
if submission[entry] == 0:
submission[entry] = 1
elif submission[entry] == 1:
submission[entry] = 2
elif submission[entry] == 2:
submission[entry] = 0
for entry in range(len(original)):
if original[entry] == 0:
original[entry] = 1
elif original[entry] == 1:
original[entry] = 2
elif original[entry] == 2:
original[entry] = 0
for ori,ensem,gold in zip(original,submission,df_gold['submission']):
print(ori,ensem,gold)
if ori != ensem:
changes += 1
if ori != gold:
ori_error += 1
if ensem != gold:
ensem_error += 1
print(changes)
print(ori_error)
print(ensem_error)
0 0 0 0 0 0 1 1 1 1 0 1 ... 20 15 13
Third model degrades performance do not include in ensemble
import pandas as pd
logit1_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/tuned_model_lr1e5_bs16_s75(5)/roberta_zero_shot/best_model/logit_results.csv')
logit2_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/scifact_model3(91.33)/roberta_zero_shot/best_model/logit_results.csv')
logit3_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/scifact_selected_ep3_1e5_16/roberta_zero_shot/best_model/logit_results.csv')
print(len(logit1_df))
print(len(logit2_df))
print(len(logit3_df))
150 150 150
import numpy as np
import ast
ensemble_logit = []
original_logit = []
model1_weight, model2_weight, model3_weight = 1.8,1.2,1
for logit_tuple1,logit_tuple2,logit_tuple3 in zip(logit1_df['logit'],logit2_df['logit'],logit3_df['logit']):
logit_tuple1 = ast.literal_eval(logit_tuple1)
logit_tuple2 = ast.literal_eval(logit_tuple2)
logit_tuple3 = ast.literal_eval(logit_tuple3)
ensemble_logit.append([model1_weight*float(logit_tuple1[0])+model2_weight*float(logit_tuple2[0])+model3_weight*float(logit_tuple3[0]),model1_weight*float(logit_tuple1[1])+model2_weight*float(logit_tuple2[1])+model3_weight*float(logit_tuple3[1]),model1_weight*float(logit_tuple1[2])+model2_weight*float(logit_tuple2[2])+model3_weight*float(logit_tuple3[2])])
original_logit.append([float(logit_tuple1[0]),float(logit_tuple1[1]),float(logit_tuple1[2])])
print(len(ensemble_logit))
ensemble_logit
150
[[-4.582047009468079, -7.972595930099487, 12.460860419273377], [-7.137682938575745, -9.142182612419129, 17.087464618682862], [8.446320748329162, 0.7103389024734499, -10.618464040756226], ...]
submission = []
original = []
for logits in ensemble_logit:
submission.append(logits.index(max(logits)))
for logits in original_logit:
original.append(logits.index(max(logits)))
changes = 0
ori_error = 0
ensem_error = 0
for entry in range(len(submission)):
if submission[entry] == 0:
submission[entry] = 1
elif submission[entry] == 1:
submission[entry] = 2
elif submission[entry] == 2:
submission[entry] = 0
for entry in range(len(original)):
if original[entry] == 0:
original[entry] = 1
elif original[entry] == 1:
original[entry] = 2
elif original[entry] == 2:
original[entry] = 0
for ori,ensem,gold in zip(original,submission,df_gold['submission']):
print(ori,ensem,gold)
if ori != ensem:
changes += 1
if ori != gold:
ori_error += 1
if ensem != gold:
ensem_error += 1
print(changes)
print(ori_error)
print(ensem_error)
0 0 0 0 0 0 1 1 1 1 1 1 ... 13 15 2
Final
np.savetxt(r'submission.txt', submission, fmt='%s')
import numpy as np
import ast
ensemble_logit = []
original_logit = []
model1_weight, model2_weight, model3_weight = 1.9,2.1,1
for logit_tuple1,logit_tuple2,logit_tuple3 in zip(logit1_df['logit'],logit2_df['logit'],logit3_df['logit']):
logit_tuple1 = ast.literal_eval(logit_tuple1)
logit_tuple2 = ast.literal_eval(logit_tuple2)
logit_tuple3 = ast.literal_eval(logit_tuple3)
ensemble_logit.append([max(model1_weight*float(logit_tuple1[0]),model2_weight*float(logit_tuple2[0]),model3_weight*float(logit_tuple3[0])),max(model1_weight*float(logit_tuple1[1]),model2_weight*float(logit_tuple2[1]),model3_weight*float(logit_tuple3[1])),max(model1_weight*float(logit_tuple1[2]),model2_weight*float(logit_tuple2[2]),model3_weight*float(logit_tuple3[2]))])
original_logit.append([float(logit_tuple1[0]),float(logit_tuple1[1]),float(logit_tuple1[2])])
print(len(ensemble_logit))
ensemble_logit
150
[[-0.5976049900054932, -2.343686580657959, 7.568054723739625], [-2.0952203273773193, -3.216470718383789, 7.667323350906372], [6.3130388259887695, 2.078376293182373, -2.7088642358779906], [6.47816801071167, -2.2090773582458496, 2.38110523223877], ...]
submission = []
original = []
for logits in ensemble_logit:
submission.append(logits.index(max(logits)))
for logits in original_logit:
original.append(logits.index(max(logits)))
changes = 0
ori_error = 0
ensem_error = 0
for entry in range(len(submission)):
if submission[entry] == 0:
submission[entry] = 1
elif submission[entry] == 1:
submission[entry] = 2
elif submission[entry] == 2:
submission[entry] = 0
for entry in range(len(original)):
if original[entry] == 0:
original[entry] = 1
elif original[entry] == 1:
original[entry] = 2
elif original[entry] == 2:
original[entry] = 0
for ori,ensem,gold in zip(original,submission,df_gold['submission']):
print(ori,ensem,gold)
if ori != ensem:
changes += 1
if ori != gold:
ori_error += 1
if ensem != gold:
ensem_error += 1
print(changes)
print(ori_error)
print(ensem_error)
0 0 0 0 0 0 1 1 1 1 1 1 ... 12 15 3
import pandas as pd
logit1_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/tuned_model_lr1e5_bs16_s75(5)/roberta_zero_shot/best_model/logit_results_phase_2.csv')
logit2_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/scifact_model3(91.33)/roberta_zero_shot/best_model/logit_results_phase_2.csv')
print(len(logit1_df))
print(len(logit2_df))
844 844
import numpy as np
import ast
ensemble_logit = []
original_logit = []
model1_weight, model2_weight = 1,1.2
for logit_tuple1,logit_tuple2 in zip(logit1_df['logit'],logit2_df['logit']):
logit_tuple1 = ast.literal_eval(logit_tuple1)
logit_tuple2 = ast.literal_eval(logit_tuple2)
ensemble_logit.append([model1_weight*float(logit_tuple1[0])+model2_weight*float(logit_tuple2[0]),model1_weight*float(logit_tuple1[1])+model2_weight*float(logit_tuple2[1]),model1_weight*float(logit_tuple1[2])+model2_weight*float(logit_tuple2[2])])
original_logit.append([float(logit_tuple1[0]),float(logit_tuple1[1]),float(logit_tuple1[2])])
print(len(ensemble_logit))
ensemble_logit
844
[[4.675937795639038, -2.1179265320301055, -3.079886865615845], [-1.6296750783920286, 3.3750624895095824, -2.8345887362957], [-0.8803869724273681, 3.936265563964844, -3.620282602310181], ...]
submission = []
original = []
for logits in ensemble_logit:
submission.append(logits.index(max(logits)))
for logits in original_logit:
original.append(logits.index(max(logits)))
changes = 0
for entry in range(len(submission)):
if submission[entry] == 0:
submission[entry] = 1
elif submission[entry] == 1:
submission[entry] = 2
elif submission[entry] == 2:
submission[entry] = 0
for entry in range(len(original)):
if original[entry] == 0:
original[entry] = 1
elif original[entry] == 1:
original[entry] = 2
elif original[entry] == 2:
original[entry] = 0
for ori,ensem in zip(original,submission):
print(ori,ensem)
if ori != ensem:
changes += 1
print()
print(changes)
1 1 2 2 1 2 2 2 1 1 ... 100
Final
np.savetxt(r'submission.txt', submission, fmt='%s')
import pandas as pd
logit1_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/scifact_sampled_model2/roberta_zero_shot/best_model/logit_results_phase_2.csv')
logit2_df = pd.read_csv('./Zero_Shot_Fact_Verification/Fact_Verification/scifact_model5/roberta_zero_shot/best_model/logit_results_phase_2.csv')
print(len(logit1_df))
print(len(logit2_df))
844 844
import numpy as np
import ast
ensemble_logit = []
original_logit = []
original_logit_2 = []
model1_weight, model2_weight = 1,1
for logit_tuple1,logit_tuple2 in zip(logit1_df['logit'],logit2_df['logit']):
logit_tuple1 = ast.literal_eval(logit_tuple1)
logit_tuple2 = ast.literal_eval(logit_tuple2)
ensemble_logit.append([model1_weight*float(logit_tuple1[0])+model2_weight*float(logit_tuple2[0]),model1_weight*float(logit_tuple1[1])+model2_weight*float(logit_tuple2[1]),model1_weight*float(logit_tuple1[2])+model2_weight*float(logit_tuple2[2])])
original_logit.append([float(logit_tuple1[0]),float(logit_tuple1[1]),float(logit_tuple1[2])])
original_logit_2.append([float(logit_tuple2[0]),float(logit_tuple2[1]),float(logit_tuple2[2])])
print(len(ensemble_logit))
ensemble_logit
844
[[6.758188962936401, -6.405615568161011, -0.04029959440231323], [-5.49228310585022, 10.1738600730896, -4.8734517097473145], [-6.30579686164856, 3.4704156816005707, 2.7231101989746094], [-2.6576818227767944, 8.672609329223633, -6.515804052352905], ...]
submission = []
original = []
original_2 = []
for logits in ensemble_logit:
submission.append(logits.index(max(logits)))
for logits in original_logit:
original.append(logits.index(max(logits)))
for logits in original_logit_2:
original_2.append(logits.index(max(logits)))
changes = 0
for entry in range(len(submission)):
if submission[entry] == 0:
submission[entry] = 1
elif submission[entry] == 1:
submission[entry] = 2
elif submission[entry] == 2:
submission[entry] = 0
for entry in range(len(original)):
if original[entry] == 0:
original[entry] = 1
elif original[entry] == 1:
original[entry] = 2
elif original[entry] == 2:
original[entry] = 0
for entry in range(len(original_2)):
if original_2[entry] == 0:
original_2[entry] = 1
elif original_2[entry] == 1:
original_2[entry] = 2
elif original_2[entry] == 2:
original_2[entry] = 0
for ori,ensem in zip(original,submission):
print(ori,ensem)
if ori != ensem:
changes += 1
print(original.count(0),original.count(1),original.count(2))
print(original_2.count(0),original_2.count(1),original_2.count(2))
print(submission.count(0),submission.count(1),submission.count(2))
print(changes)
... 1 1 0 0 0 0 2 2 2 0 2 2 0 0 2 2 2 2 0 0 1 1 0 0 1 2 0 0 2 2 0 1 2 1 0 0 2 2 2 1 0 0 200 209 435 328 204 312 308 204 332 186
Final results, a maximum of 228 changes, weights tuned to allow 186
np.savetxt(r'submission.txt', submission, fmt='%s')